mne.read_epochs#
- mne.read_epochs(fname, proj=True, preload=True, verbose=None) EpochsFIF[source]#
Read epochs from a fif file.
- Parameters:
- fnamepath-like | file-like
The epochs to load. If a filename, should end with
-epo.fifor-epo.fif.gz. If a file-like object, preloading must be used.- projbool | ‘delayed’
Apply SSP projection vectors. If proj is ‘delayed’ and reject is not None the single epochs will be projected before the rejection decision, but used in unprojected state if they are kept. This way deciding which projection vectors are good can be postponed to the evoked stage without resulting in lower epoch counts and without producing results different from early SSP application given comparable parameters. Note that in this case baselining, detrending and temporal decimation will be postponed. If proj is False no projections will be applied which is the recommended value if SSPs are not used for cleaning the data.
- preloadbool
If True, read all epochs from disk immediately. If
False, epochs will be read on demand.- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- Returns:
- epochsinstance of
Epochs The epochs.
- epochsinstance of
Examples using mne.read_epochs#
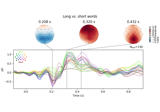
Visualising statistical significance thresholds on EEG data
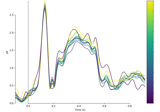
Analysing continuous features with binning and regression in sensor space